Phenotypes and Diseases
Rare diseases
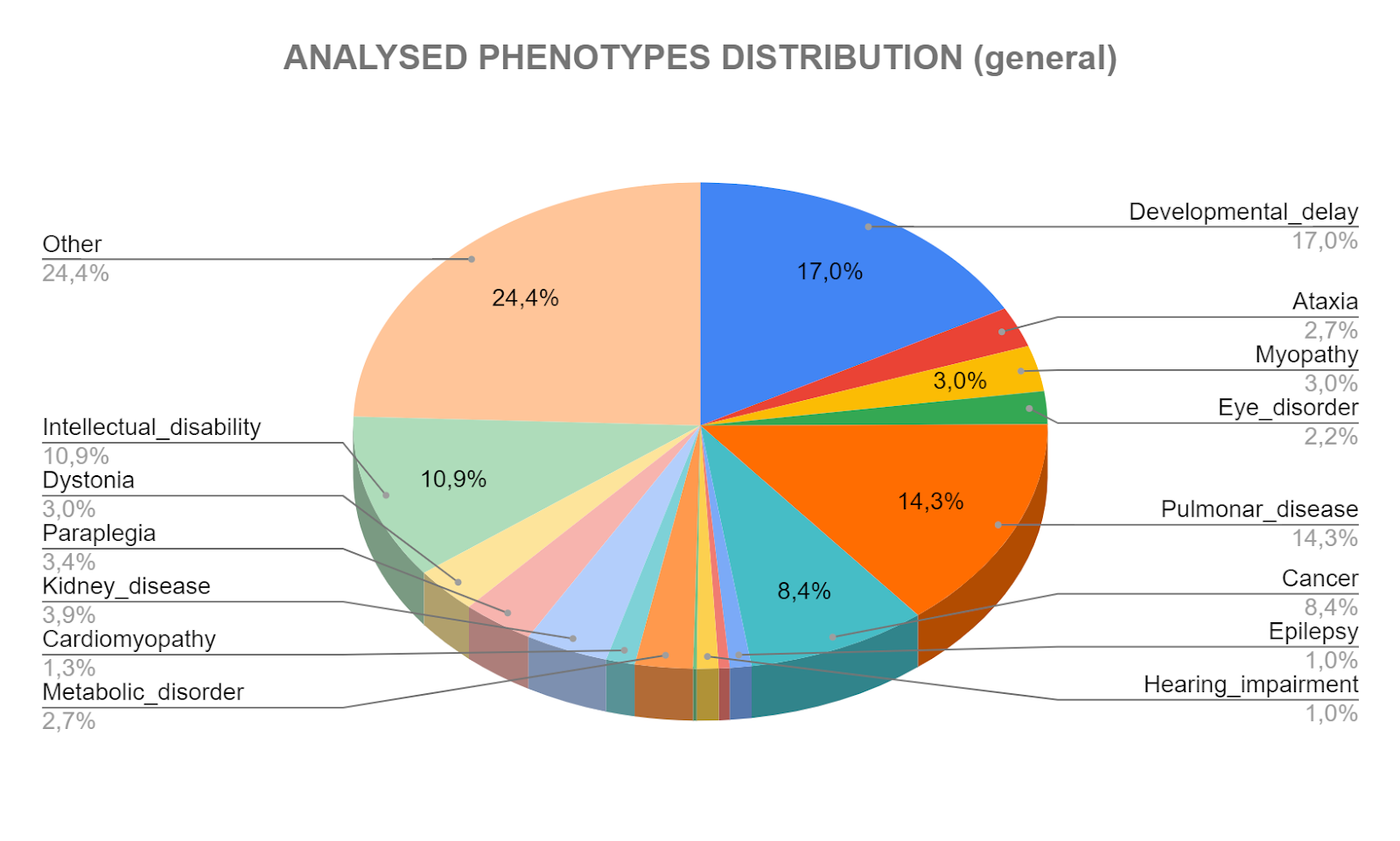
A Rare Disease (RD) is defined in the European Union as those with a prevalence of less than one among 2000. Although any individual RD has a very low prevalence, there are between 6000 and 7000 RDs, which results in a collective prevalence similar to the observed in many common diseases, constituting a substantial health problem. It is difficult to have a real assessment of the number of people affected by an RD, but conservative estimations point to about 3 million people affected only in Spain, which represents a 6% of the population. Despite the molecular cause is known for a significant number of RDs, there are specific treatments available only for a small fraction of them (less than 5%).
One of the main problems of RD is the lack of an effective treatment due to the absence of a proper diagnosis, among other factors. As previously mentioned, RDs as a whole are a serious health problem, being, most of them, remarkably detrimental for patient life quality, since many RDs present complications or comorbidities derived from the lack of treatment, which constitutes an extra socioeconomic burden. Additional to this is the lack of independence of the patient, which causes psychologic prejudices and prevents his/her career development. Moreover, many RD patients experience a delay in their diagnosis, that can last months, years or even their entire life (12 years on average), with 40% of patients having a wrong diagnosis or remaining undiagnosed, according to EURORDIS. Most families identify this delay as one of the main concerns related to its struggle with the disease. Not knowing what is going on and how to deal with it adds a weight to the burden that families have to bear.
Given that an adequate diagnosis and treatment, allows patients to arrive to have a considerable improvement in their life quality and that of their families, we, as a clinical community, have the duty to offer a diagnosis in the most accurate and fast way possible, making use of all available tools, including NGS and its analysis.








