This workflow describes the entire procedure followed to determine and analyse somatic variants from raw sequencing data. The provided pipeline is based on the GATK best practices and their corresponding tutorials (updated to GATK v4.1.2)
Graphical diagram
Diagram available in: somatic_workflow_diagram.odp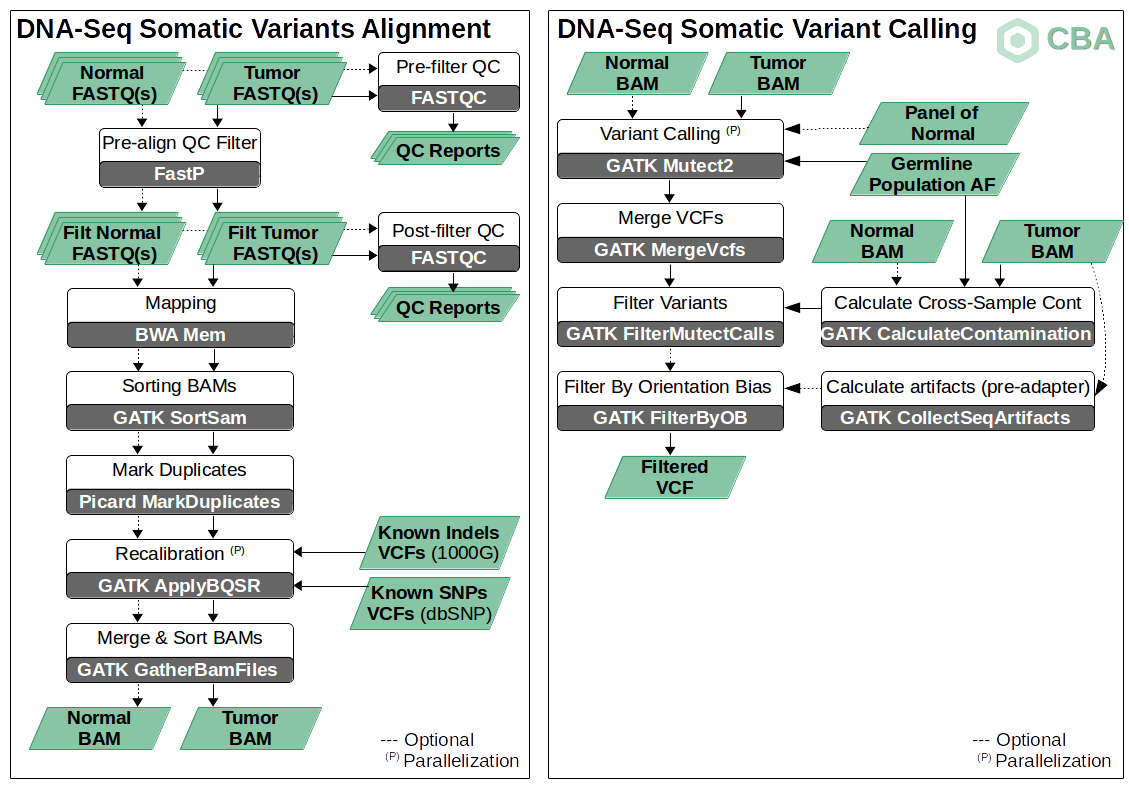
General description
The purpose of the somatic variant pipeline is to call alterations that are identified when contrasting two samples from the same individual, usually one tumor sample against one normal sample. This way those variants being specific from the tumor sample (somatic variants) against the normal sample as well as different to the reference are called.
According to the GATK recommendations (see Diagram above), both tumor and normal samples can be initially treated as separated samples in the alignment phase, thus following a similar procedure as standard germline pipelines (Pre-processing & QC, Mapping and Recalibration & Post-processing). After these steps, GATK Mutect2 tool is applied for somatic variant calling. This caller can work with a tumor-normal pair of aligned samples (BAM files) but it can also accept one single tumor sample (tumor-only mode, see below for details). Additionally, several resources like collections of allele frequencies by populations or Panel of Normals (PoN) are considered to improve the accuracy in the calling of somatic mutations.
This pipeline has been developed by using the purpose-specific workflow language WDL (Workflow Description Language). The implemented WDL scripts are specifically designed to be executed in our internal cluster by using the orchestration tool Cromwell from Broad Institute. The entire pipeline is privately available for the member of our center as a GitLab repository. There are two different versions currently in production for this pipeline:
v1.0 -> Initial pipeline version. Corresponding with GATK v4.0.9 best practices. v1.1 -> Pipeline updated for GATK v4.1.2 new features and improvements.
Tumor-only mode
As shown in the above diagram, our somatic variant pipeline allows to be executed by using tumor-normal pairs as well as only tumor samples (tumor-only mode). This mode usually carries several conditions, cautions and/or limitations. Some of the main recommendations to take into account are:
-
Do not using a matched normal sample usually provides one more order of magnitude in terms of number of variants than tumor-normal pairs. This meaning that the tumor-only mode results in a more sensitive but less specific solution (higher false-positive rate). Nevertheless, this problem can be reduced later by an accurate annotation of the variants.
-
It is strongly advisable to apply of a Panel of Normal (PoN) when working in tumor-only mode. The usage of a PoN avoids the leaking of common germline variants into the results but also (and most important) other artifacts and systematic noisy variants. For specific details about how to create an accurate PoN, please see the following section.
-
Additional external resources including population frequencies of common germline variants or other common variants produced by contamination are also recommendable to decrease the number of errors and potentially germline variants in final results. Some of these recommended resources can be:
Germline Population AFs -> gs://gatk-best-practices/somatic-b37/af-only-gnomad.raw.sites.vcf Contamination information -> small_exac_common_3_b37.vcf.gz
Creation of Panel of Normal (PoN)
As briefly mentioned above, adding a Panel of Normal (PoN) is helpful to avoid false positive variants and additional artifacts. The PoN is specially relevant for reinforce the calling step when normal samples are not available (tumor-only mode). Our pipeline has followed some of the well-described tutorials by GATK about creation and application of PoNs:
Somatic Variants is not only a comparison
When using a PoN, GATK includes some additional FILTER tags to describe variants specifically found by the PoN:
-
If the variant identically matches one variant reported in the PoN, the variant is not called.
-
If the variant matches the position of another variant reported in the PoN, but there is not an identical match in the alleles (allele-mismatch) the variant is tagged as panel_of_normals. The field IN_PON in the FILTER column of the VCF file includes the identified allele differences.
