This page describes the entire workflow and the different configurations of the RNA-Seq pipeline developed at the Clinical Bionformatics Area (CBA). The pipelines has been designed to be versatile and adaptive depending on the demanding features and available resources from the final user.
Graphical Diagram
Diagram also available in PDF: cbra_rnaseq_pipeline_diagram.pdf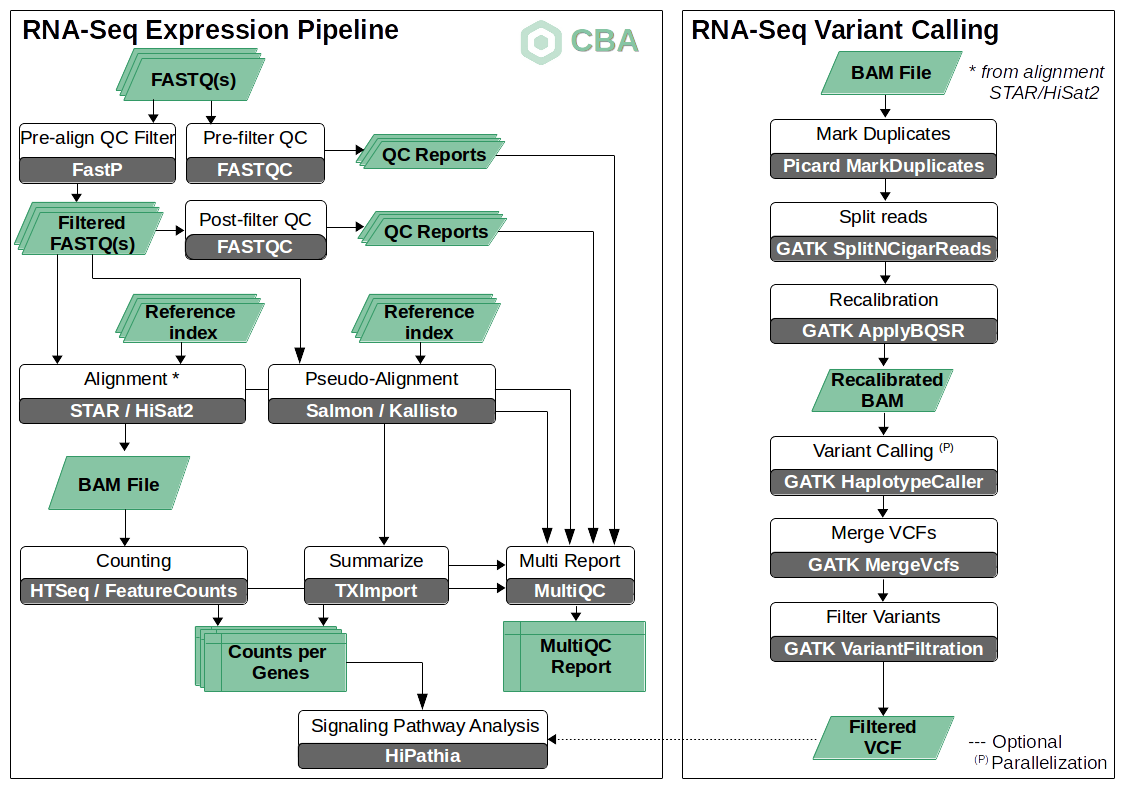
General Description
The main purpose of the RNA-Seq pipeline is the quantification and analysis of differential expression based on data from sequencing technologies. Commonly, RNA-Seq analyses allow to count (quantify) the number of reads aligned in each region of the reference, thus having a precise estimation about how much each gene is expressed (after a careful normalization).
This pipeline includes the standard steps for a common RNA-Seq analysis: alignment, pre- and post-alignment QC and filtering, normalization and read counts. Additionally, the pipeline is enriched with two more specific steps:
Post-processing signaling pathway analysis based on expression results using our in-site developed HiPathia tool.
An optional variant calling procedure using RNA-Seq reads according to the GATK best practices for variant calling on RNA-Seq.
This pipeline was developed with the strong objective of making it flexible and versatile. In this sense, the implemented pipeline allows users configuring their own options according to their requirements and computational resources. For instance, alternative paths can be selected for the alignment step: running full alignment strategies like STAR (v2.7.2b) or HiSat2 (v2.1.0); choosing other pseudo-alignment solutions like Salmon (v0.13.0) or Kallisto (v0.46.0). In general, full alignments are generally more computationally demanding but more accurate. Also, users can decide whether they want to add a variant calling analysis or not. Please, note that variant calling is only possible when we have an aligned read file (BAM files) from alignment steps (full alignment strategies).
Similarly to other pipelines in CBA, the RNA-Seq pipeline has been developed by using the purpose-specific workflow language WDL (Workflow Description Language). All the steps of the pipeline have been wrapped into WDL tasks. In addition, each task has been implemented to be run on an independent unit of containerized software using docker. Also, the orchestration tool Cromwell from Broad Institute has been used for execution purposes.
